About
Reproducibility is a key component of science, as the replication of findings is the process by which they become knowledge. It is widely considered that many fields of science, including medical imaging, are undergoing a reproducibility crisis. This has led to the publications of various guidelines to improve research reproducibility, such as the checklist proposed by MICCAI. On top of a clear description of the materials, methods and results, the checklist includes several items related to the code, which correspond to the elements of the ML Code Completeness Checklist in use at NeurIPS. If such checklists constitute useful guides, some users may need further instructions to actually comply with them. This tutorial aims to help MICCAI members adopt good machine learning research practices to enable reproducibility.
Learning objectives of the tutorial include:
- An understanding of the different meanings of reproducibility;
- Raising awareness and improving participants’ ability to detect possible sources of irreproducibility in scientific articles;
- A good knowledge of the main data management principles and of the best coding and machine learning research practices;
- A clear view of the tools that can be used to perform reproducible research.
Keynote

Dr. Koustuv Sinha
Meta AI New York - Fundamental AI ResearchKoustuv Sinha is a Research Scientist at Meta AI New York, in the Fundamental AI Research (FAIR) team. He did his PhD from McGill University (School of Computer Science) and Mila (Quebec AI Institute), supervised by Joelle Pineau, in the wonderful city of Montreal, QC, Canada. He spent a significant portion of his PhD being a Research Intern (STE) at Meta AI (FAIR), Montreal.
He now is an associate editor of ReScience C, a peer reviewed journal promoting reproducible research, and he is the lead organizer of the annual Machine Learning Reproducibility Challenge. His work has been covered by several news outlets in the past, including Nature, VentureBeat, InfoQ, DailyMail and Hindustan Times.
Program
| 8.00 | - | 8.10 | Introduction |
| 8.10 | - | 8.55 | Keynote Koustuv Sinha (Meta AI New York) |
| 8.55 | - | 9.15 | Setup explanations |
| We will take some time to ensure has the correct environment for the second exercise. Please refer to the corresponding section to start setting up your environment. | |||
| 9.15 | - | 10.00 | Presentation of the MICCAI reproducibility checklist: practical tips to achieve reproducibility |
| 10.00 | - | 10.30 | ☕ Coffee Break |
| 10.30 | - | 11.45 | Critical review tutorial (in teams) |
| During this tutorial, participants will be teamed up to conduct critical reviews of pairs of (fake) papers and GitHub repos all performing the same task. | |||
| 11.45 | - | 12.15 | Reproducibility tutorial (solo) |
| During this last part, the participants will have access to a paper and the corresponding repo which meets all the requirements of the MICCAI reproducibility checklist. | |||
| 12.15 | - | 12.30 | Wrapping up |
Exercise 1 - Critical review tutorial
The goal of this first exercise is to fill an adapted version of the MICCAI reproducibility checklist for different fake papers.
Content of the papers
All the papers have the same content: their objective is to train a deep learning network to perform a binary classification, Alzheimer's disease patients VS cognitively normal participants, using brain T1-MRI. Then they explain their algorithm using an explainability method generating an attribution map which is showing what the network uses in the image to take a decision.
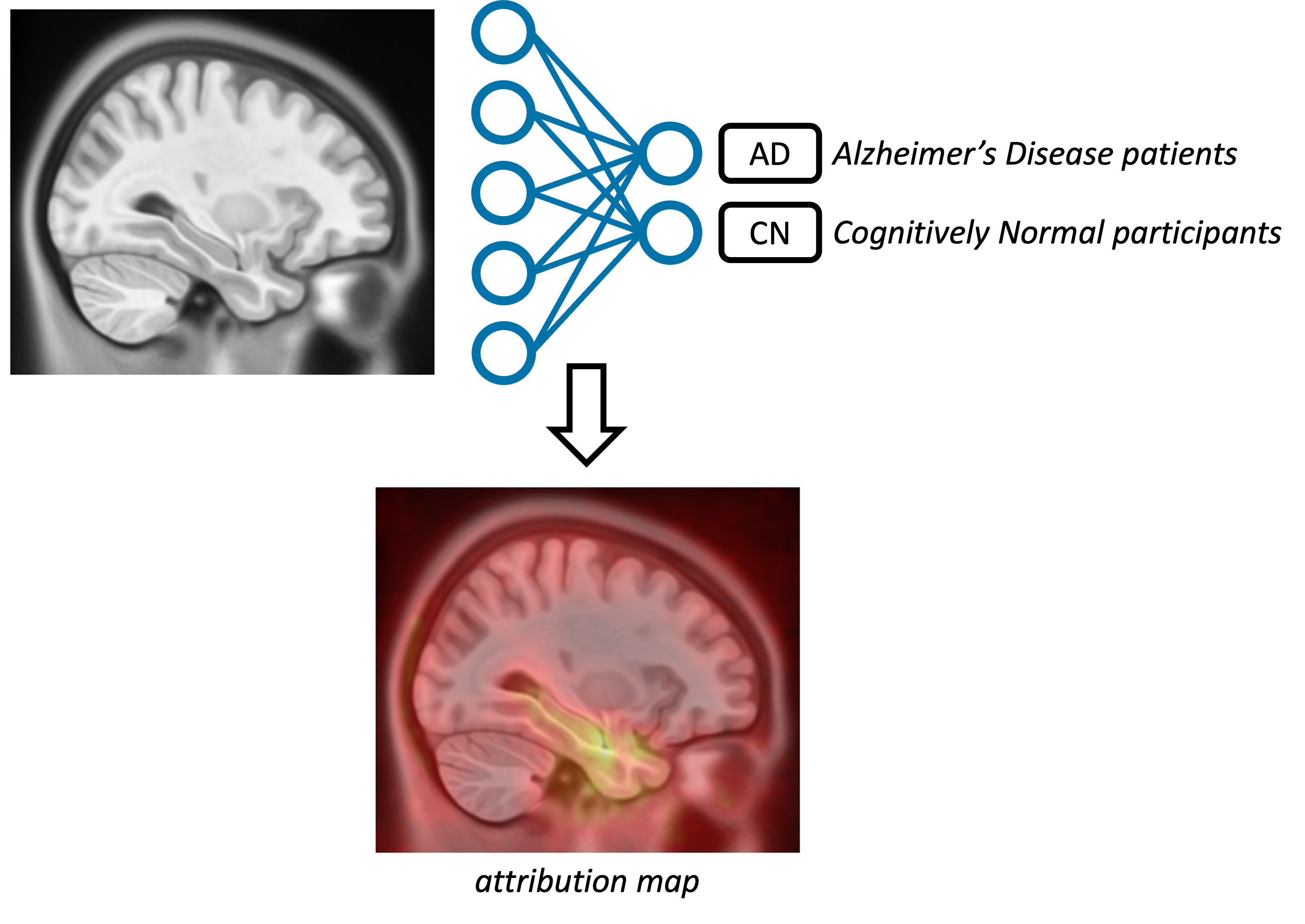
They also all have the same organisation. The abstract and introduction sections are all identical and do not include any useful information, except the link to the corresponding GitHub repo at the end of the introduction.
Content of the checklist
For all papers, the participants have to fill the same checklist. Each section of the checklist is related to one or more sections of the article or the GitHub repo.
-
Data set - Sections 2 + 3.1, GitHub repo
- Availability - Open-source / Semi-public / On request / Not findable
- Reference (citation / link) - Yes / No
- Definition of the labels (AD & CN) - Not described ↔ Reproducible
- Description of the images (acquisition protocol/preprocessing) - Not described ↔ Reproducible
-
Code - Sections 3.2 + 3.3, GitHub repo
- Availability - Open-source / Semi-public / On request / Not findable
- Dependencies - Fixed versions / No versioning / None
- Preprocessing code - None ↔ All experiments
- Training code - None ↔ All experiments
- Evaluation code - None ↔ All experiments
- Explainability code - None ↔ All experiments
- Pre-trained models - Open-source / Semi-public / On request / None
- Minimal list of commands to run the code - None ↔ All experiments
-
Computational setup - Section 3.2
- Runtime - Yes / No / Not applicable
- Memory footprint - Yes / No / Not applicable
- Computing infrastructure - Yes / No / Not applicable
-
Training - Section 3.2
- Hyperparameter search - None ↔ All experiments
- Hyperparameter values - None ↔ All experiments
-
Evaluation - Sections 3.3 + 4
- Validation procedure - Not described ↔ Reproducible
- Error bars / confidence intervals - None ↔ All experiments
- Statistical significance - None ↔ All experiments
Links to the materials
The papers, codes and checklists can be found here:
Exercise 2 - Reproduce It Yourself!
The goal of this second exercise is to replicate an experiment from a research paper that adheres to reproducibility standards. The intention is to demonstrate how straightforward this process can be when following a reproducibility checklist. This step is crucial as it illustrates how a well-documented and reproducible methodology can greatly simplify the replication of research findings, thereby enhancing the verifiability and reliability of scientific work.
This time, you do not have to fill out a form, but you have to run the code (which should be really easy 😉). The final goal is to reproduce the tables and figures expliciting the results in the paper:
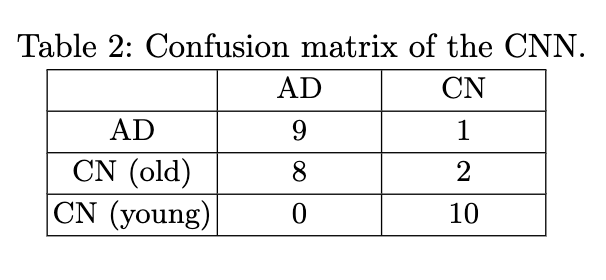
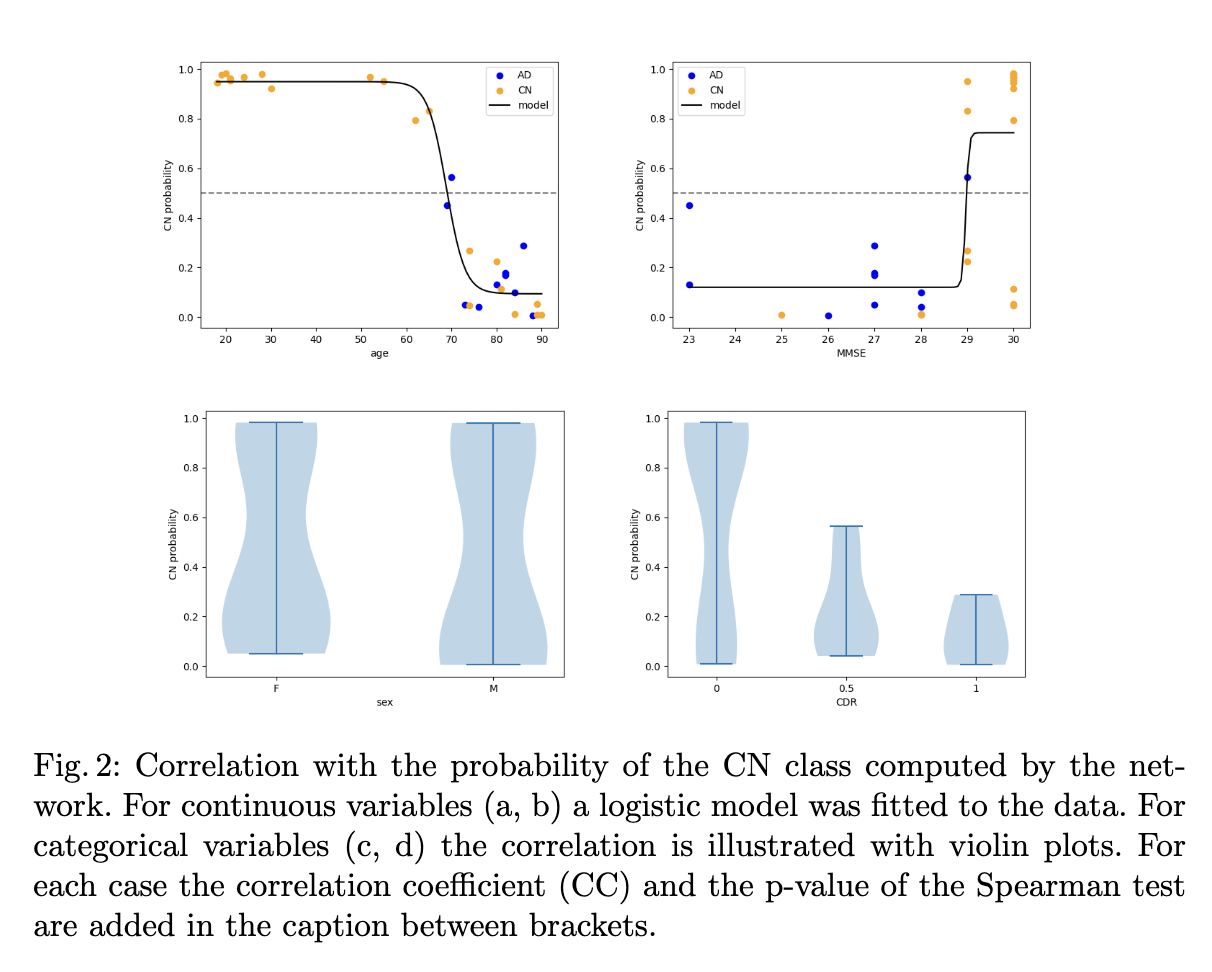
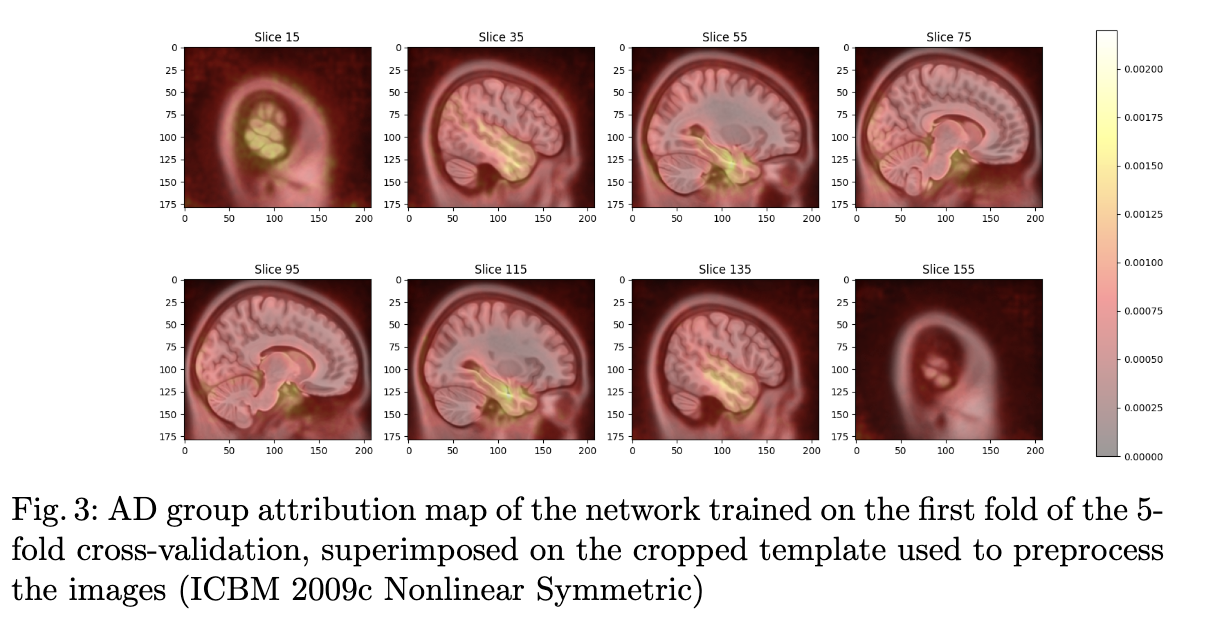
Content of the paper
The objective of the paper is to explore the transferability of a CNN trained on the ADNI dataset to the OASIS-1 dataset for Alzheimer's disease detection. This paper is applying a pre-trained network to a data set. We decided to make attendees train the network to limit the need for computational resources.
Preparation steps
To limit download time, we redistributed the part of the OASIS data set we will be using here.
Please also make sure that you have Anaconda or Miniconda installed, as this tutorial will ask you to create a conda environment.
Organizers
This reproducibility tutorial is organized by Ninon Burgos and Elina Thibeau-Sutre. You can reach us via e-mail.
Dr. Ninon Burgos (CNRS - Paris Brain Institute - ARAMIS Lab) is interested in the processing and analysis of neuroimages, mainly image synthesis, on the use of images to guide diagnosis, and on the application of these methods to the clinic.
Camille Brianceau (Paris Brain Institute - ARAMIS Lab) is the main developer of the ClinicaDL library, a project meant to enhance reproducibility in studies using deep learning applied to neuroimaging.
Dr. Elina Thibeau-Sutre (Mathematics of Imaging and AI - University of Twente) is interested in the robustness and interpretability of deep learning applied to medical problems such as colorectal cancer surgery (project awarded by CMI-NEN) and subdural brain hematoma treatment (project awarded by PIHC 2023).

Dr. Ninon Burgos
CNRS - Paris Brain institute ARAMIS Lab
Camille Brianceau
Paris Brain institute ARAMIS Lab


